Original paper: A BERT model generates diagnostically relevant semantic embeddings from pathology synopses with active learning
Introduction
Pathology synopses are a crucial aspect of the diagnostic process. They involve experts observing human tissue and summarizing their findings in semi-structured or unstructured text. These synopses are critical in helping physicians make accurate diagnoses and develop appropriate treatment plans.
However, the limited number of specialists available to interpret pathology synopses can be a significant barrier to accessing this essential information. Fortunately, deep learning offers a solution to this problem.
Main Problems
How to build the pathology synopses dataset, as only limited number of experts can interpret these synopses?
How to extract info from pathology synopses, which are complex, and require a high degree of domain-specific knowledge to interpret.
Solutions Applied
The overall modeling process:
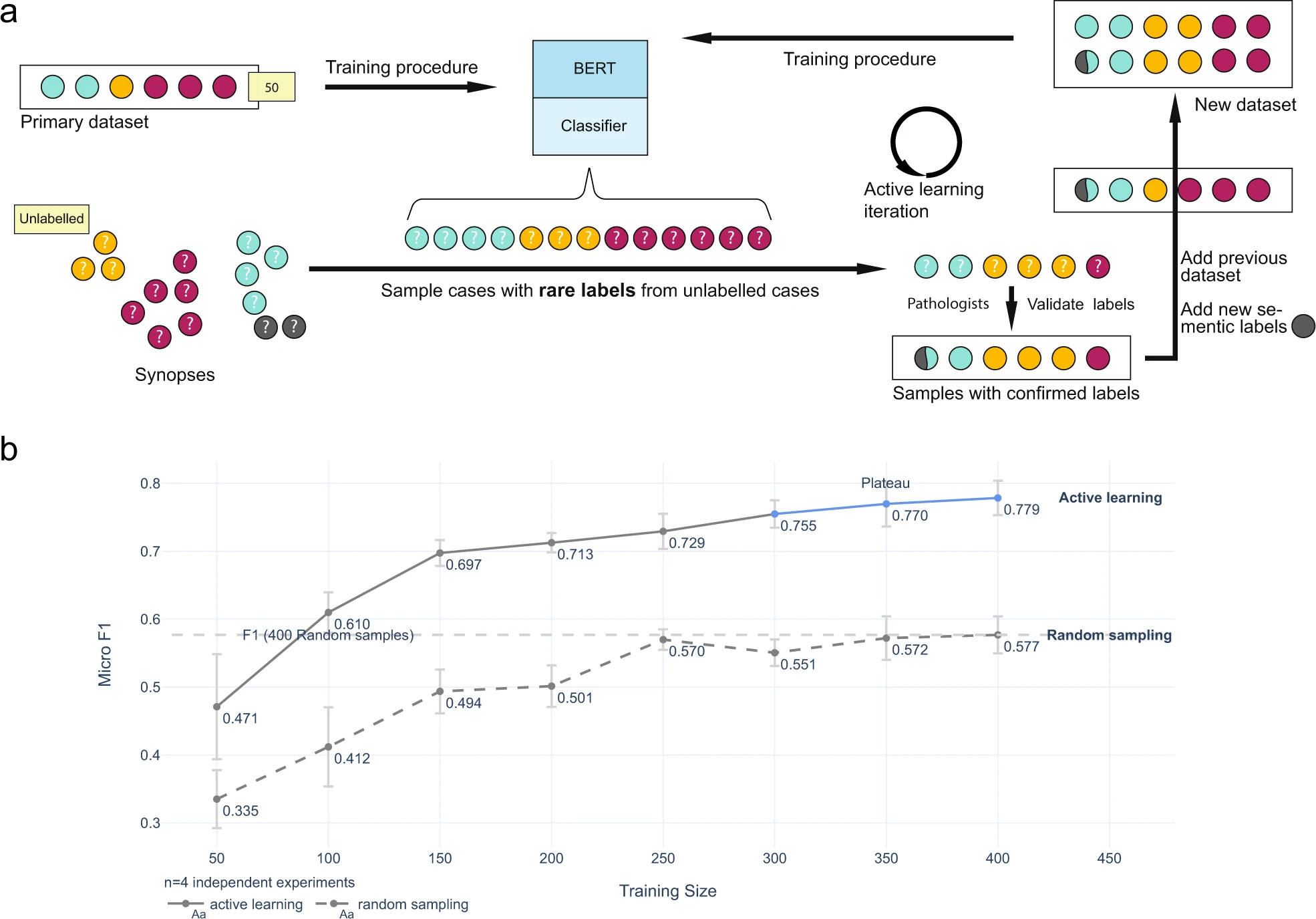
Active learning for pathology synopses dataset
We used an active learning approach was used to develop a set of semantic labels for bone marrow aspirate pathology synopses.
In active learning, the machine learning model is not just trained on a fixed dataset, but it is also allowed to query a human expert for labels for selected examples. Here, we start with a small set of labeled examples. These labeled examples would be used to train a machine learning model to map the pathology synopses to semantic labels. The model would then be used to generate predictions for a larger set of unlabeled examples.
Rather than simply accepting the model’s predictions, the active learning approach selected a subset of the unlabeled examples that the model is least certain about and present them to a human expert for labeling. By doing this, the machine learning model can learn from the expert’s input and improve its accuracy over time.
This process is repeated iteratively, with the machine learning model being updated with the newly labeled examples, and the model making new predictions on the remaining unlabeled examples. This approach significantly reduced the number of labeled examples required to develop an accurate set of semantic labels for pathology synopses, which is essential as it can be time-consuming and expensive to obtain large amounts of labeled data.
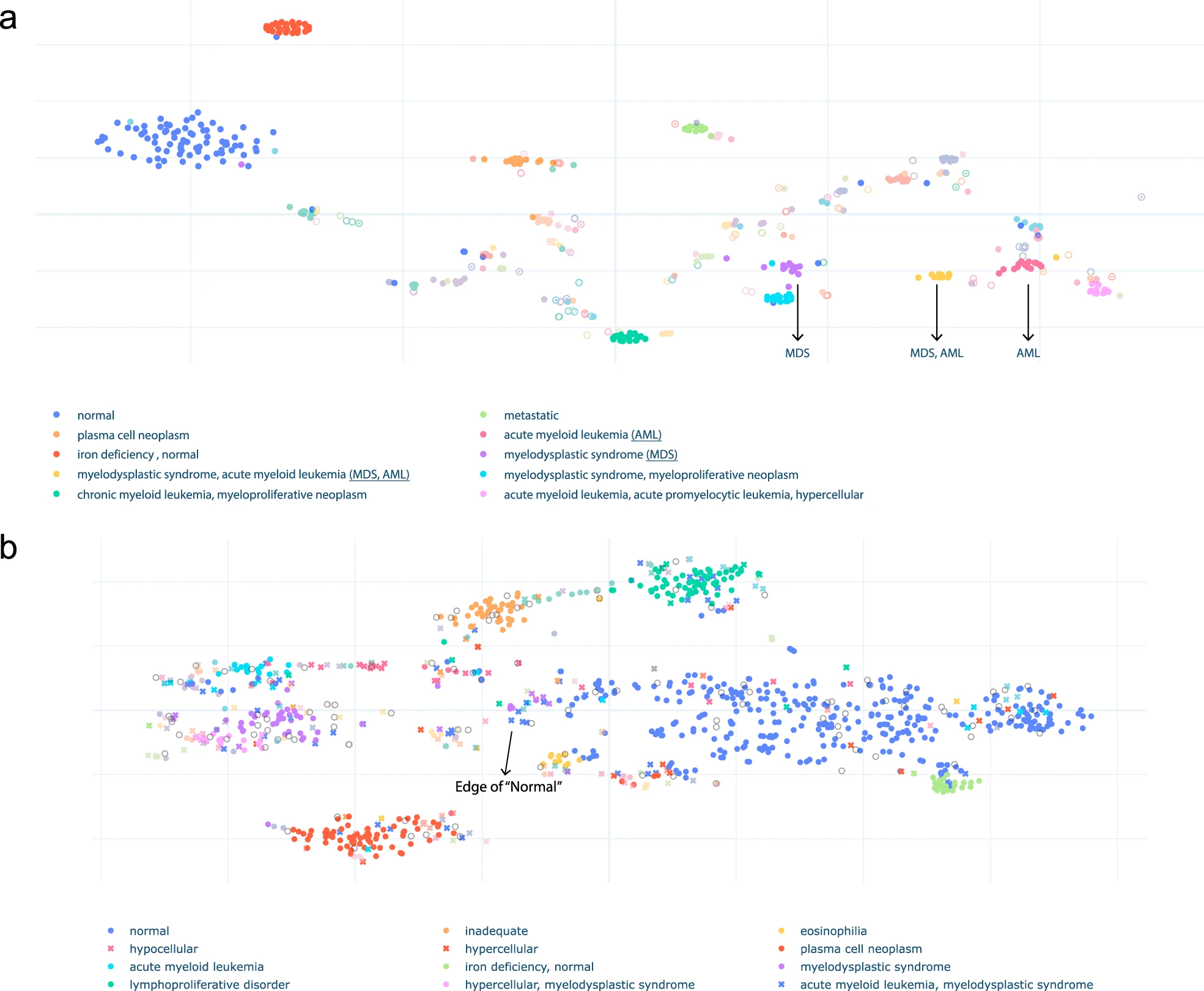
The active learning process and its result:
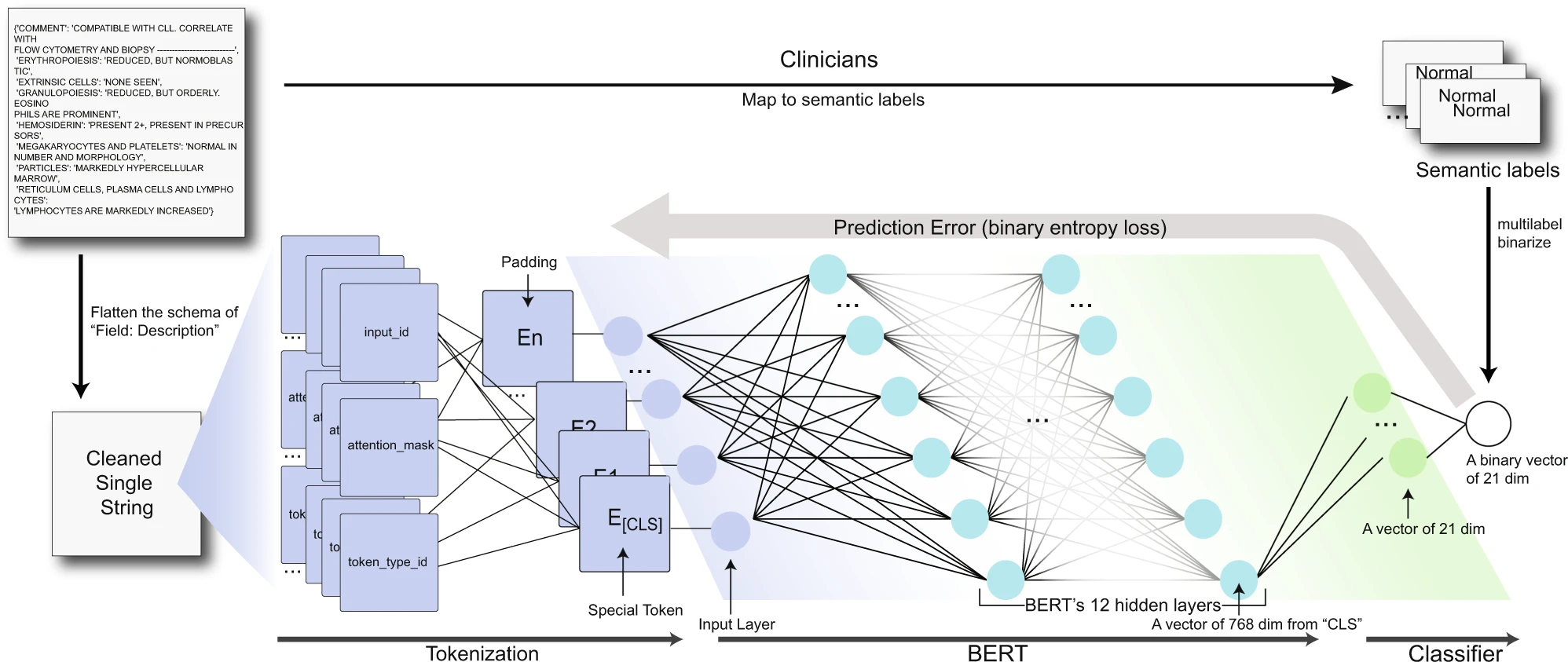
Transformer for pathology synopses
A transformer-based deep-learning model (BERT) was then trained to map these synopses to one or more semantic labels and to extract learned embeddings from the model’s hidden layer.
BERT (Bidirectional Encoder Representations from Transformers) is a state-of-the-art natural language processing (NLP) model that can generate meaningful embeddings from unstructured texts. BERT is based on a self-attention mechanism that allows it to process input sequences in a way that captures the relationships between all of the input tokens. When BERT processes a pathology synopsis, it generates a sequence of hidden states that capture the meaning of each token in the input. These hidden states are then used to generate a fixed-length vector, called an embedding, that represents the meaning of the entire input sequence.
The key advantage of BERT over traditional NLP models is that it is pre-trained on large amounts of text data using a masked language modeling task. This pre-training allows BERT to learn general language representations that can be fine-tuned for a specific task, such as mapping pathology synopses to semantic labels.
To generate meaningful embeddings from unstructured pathology synopses, BERT is fine-tuned on the labeled dataset of pathology synopses created by active learning. During fine-tuning, the model learns to map the unstructured text input to the correct semantic labels by adjusting the weights of its neural network based on the labeled examples.
The learned embeddings generated by BERT capture the semantic meaning of the input sequence in a way that is useful for downstream tasks such as diagnostic classification. By leveraging the power of deep learning and pre-training, BERT can generate embeddings that capture the subtle nuances and context-specific information present in pathology synopses, ultimately leading to more accurate diagnoses and improved patient outcomes.
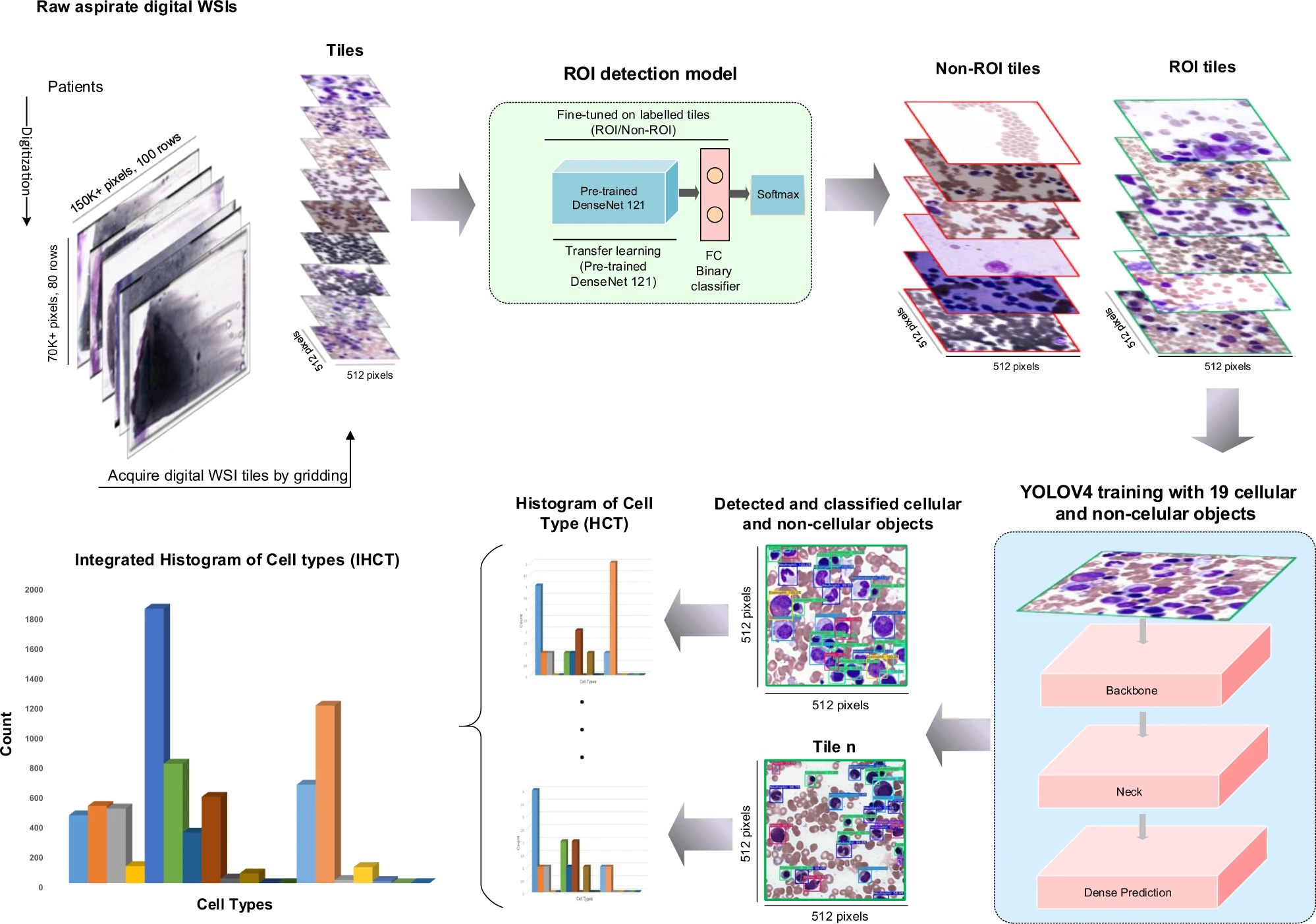
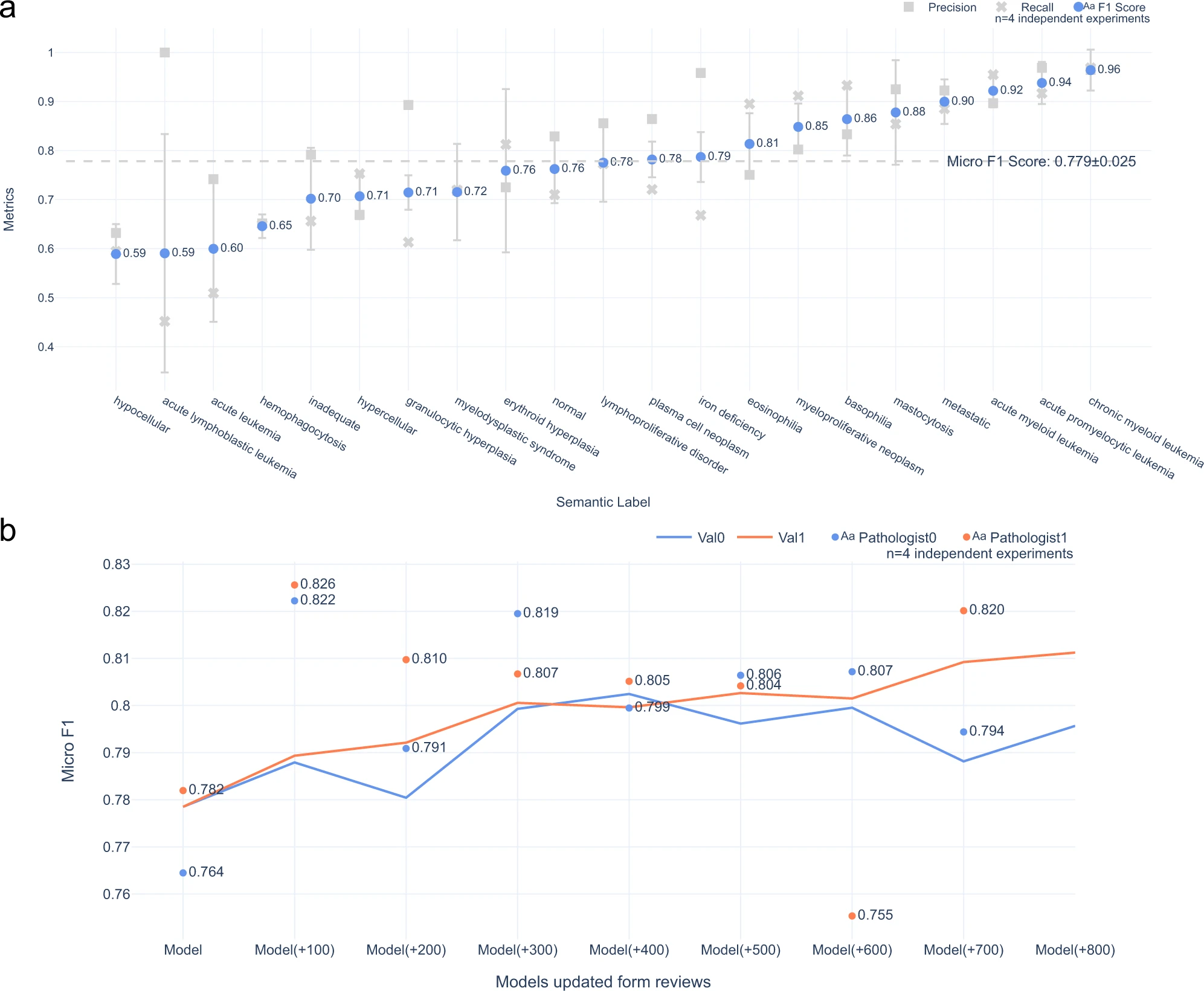
Model performance in embedding extraction:
Results
The results of the study demonstrated that with a small amount of training data, the transformer-based natural language model was able to extract embeddings from pathology synopses that captured diagnostically relevant information. On average, these embeddings could be used to generate semantic labels mapping patients to probable diagnostic groups with a micro-average F1 score of 0.779 ± 0.025.
Pathology synopses play a critical role in the diagnostic process, and their interpretation requires a high degree of domain-specific knowledge. The success of this study provides a generalizable deep learning model and approach that can unlock the semantic information inherent in pathology synopses. By using transformer-based deep-learning models to extract meaningful embeddings from pathology synopses, we can unlock the semantic information inherent in these synopses and improve the accuracy and speed of diagnoses.
Model performance in label prediction:
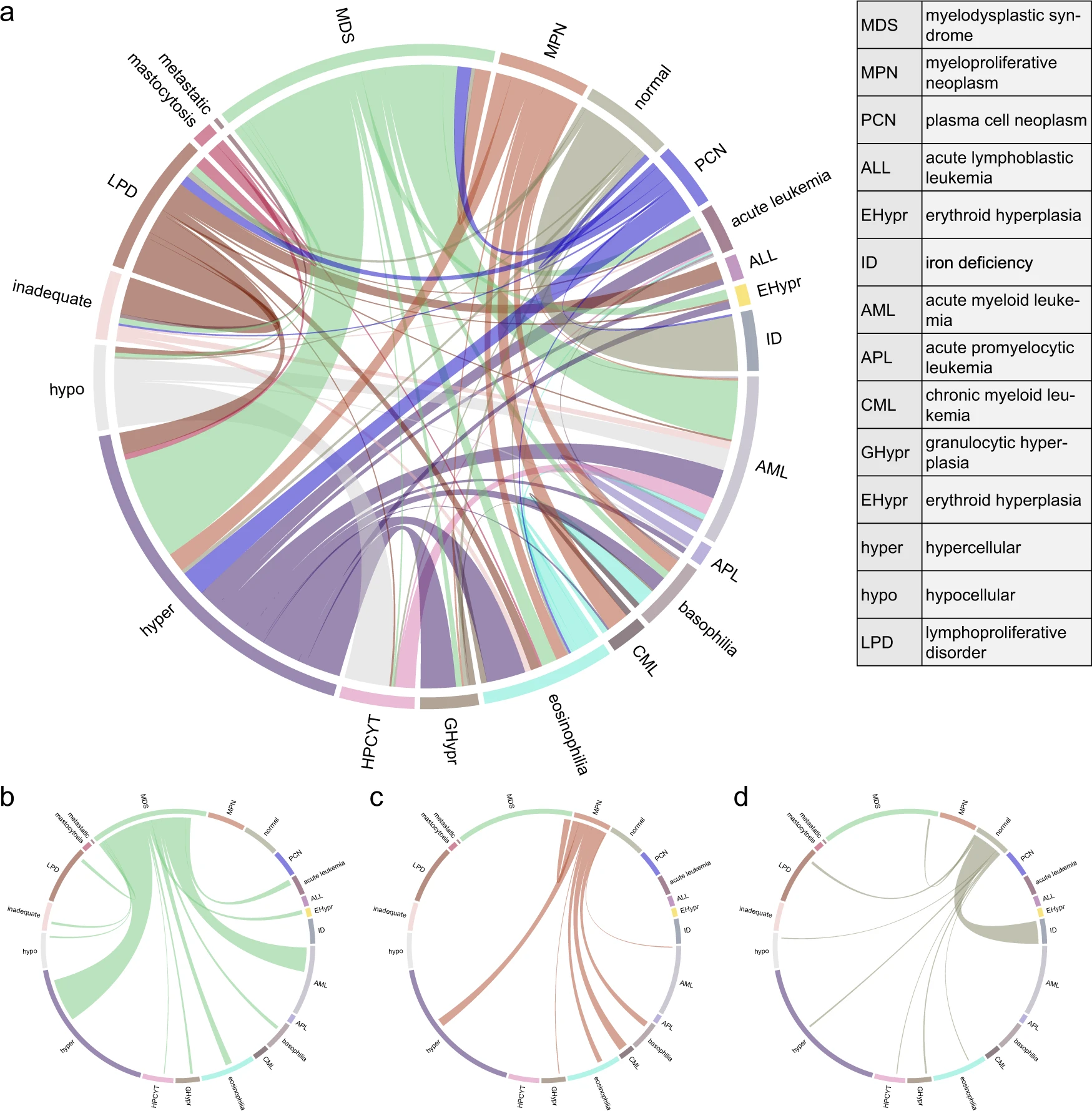
Co-occurrence of the predicted labels: