Introduction
In medical research, histology provides essential information for tumor diagnosis and treatments. Anatomic pathologists can classify disease progression by evaluating histological characteristics, such as nuclear atypia, mitotic activity, cell density, and tissue structure.
With the advancement of gene sequencing, it is now possible to directly observe the genome’s biomarkers, such as gene expression and epigenetic modification changes, to analyze tumor symptoms. However, histological analysis is still an essential and indispensable tool for predicting the future course of patients. Histological characteristics show the overall impact of molecular-level changes. Meanwhile, the tissue can provide intuitive visual information to help medical staff judge cancer’s invasiveness. However, histological analysis has significant flaws — that is, it is highly subjective and hard to repeat to obtain the same results.
In contrast, for computers, the results are derived from a specific series of computations. When the calculation process is unchanged, the same data input will produce the same result.
Also, computers can extract all the histological imaging information, while people can only observe specific areas empirically. In this way, computers can use the neglected data in histological imaging to make the diagnosis more comprehensive.
Therefore, the analysis of histological imaging by computer can overcome the shortcomings of manual tissue analysis and extract information ignored by experts. As a result, the application of computer analysis of histological imaging to assist medical diagnosis is very promising.
However, in most recent studies, experts must label the useful areas of pathological images (Region of Interest). Also, the models they presented are large and complex. Consequently, the models are not easy to use on an ordinary personal computer, limiting their usefulness.
Main Problems
How to build a model that can provide “time-event” predictions by analyzing breast cancer histological imaging?
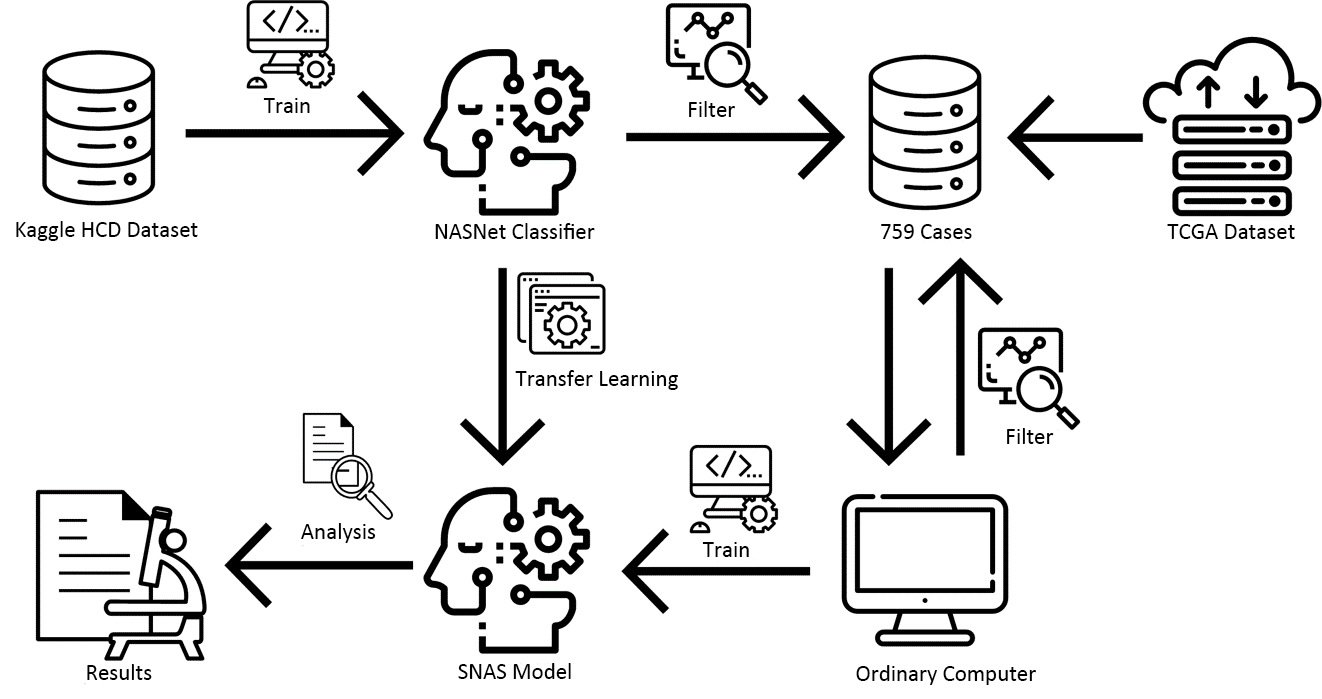
How to set up a fully automatic “end-to-end” batch processing system, which takes .svs pathological images as input, and survival models as output?
How to make this model be trained and used on ordinary computers?
Solutions I applied
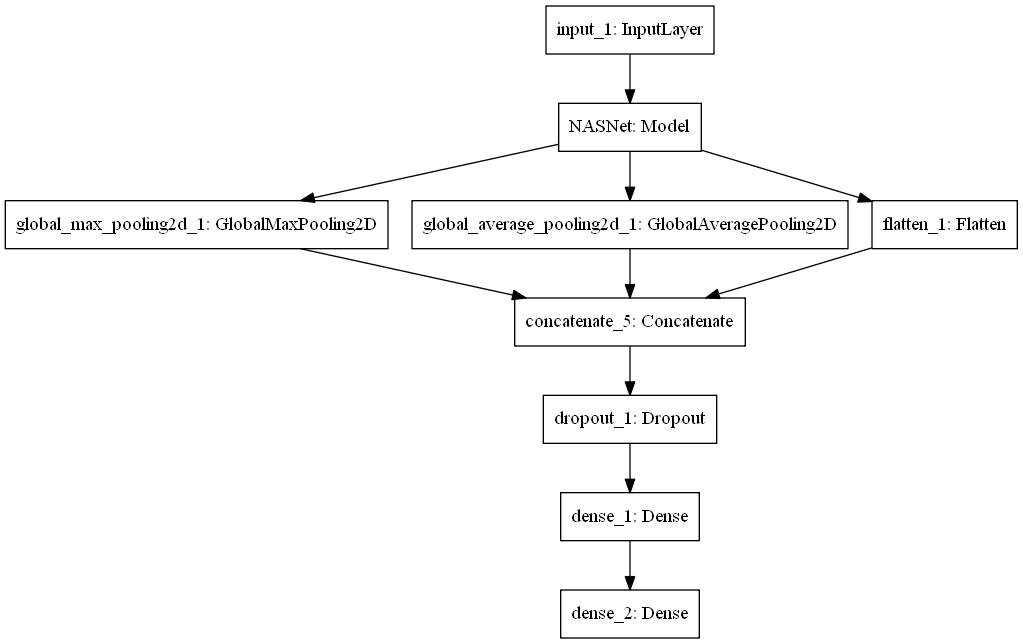
In May 2017, Google Brain designed the AutoML, which can generate artificial intelligence (AI) models. NASNet is a neural network model that Google researchers use reinforcement learning and AutoML as a controller automatically trained. In 2018, NASNet was already the best model in the field of image recognition. So I choose NASNet as the base model to built my Breast Cancer Survival Neural Network Model (SNAS) like below.
The raw model.
Put genome data into the model, and form the one below.
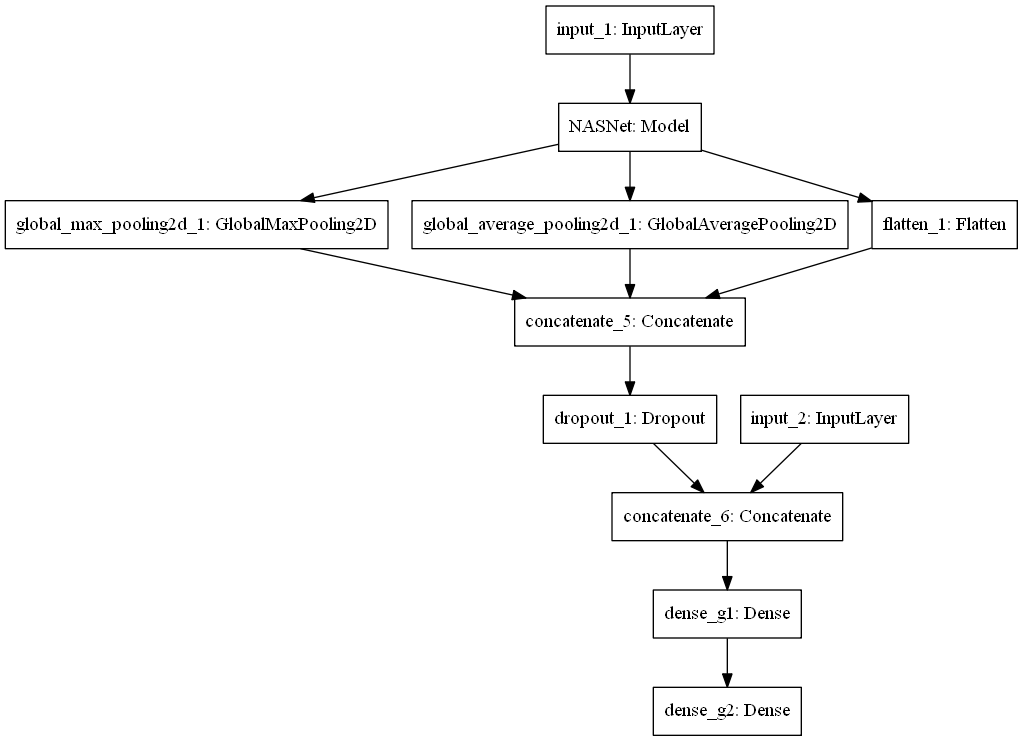
NASNet model has two types, NASNet large model and NASNet mobile model. NASNet mobile is one of the smaller models, and its hardware requirements are smaller, but it can still achieve powerful image recognition functions. To make this model be trained and used on ordinary computers, NASNet mobile model was selected.
A system including three parts: segmentation, extraction, and training, is designed to achieve “end-to-end” batch processing. Every piece does not require human intervention.
Results
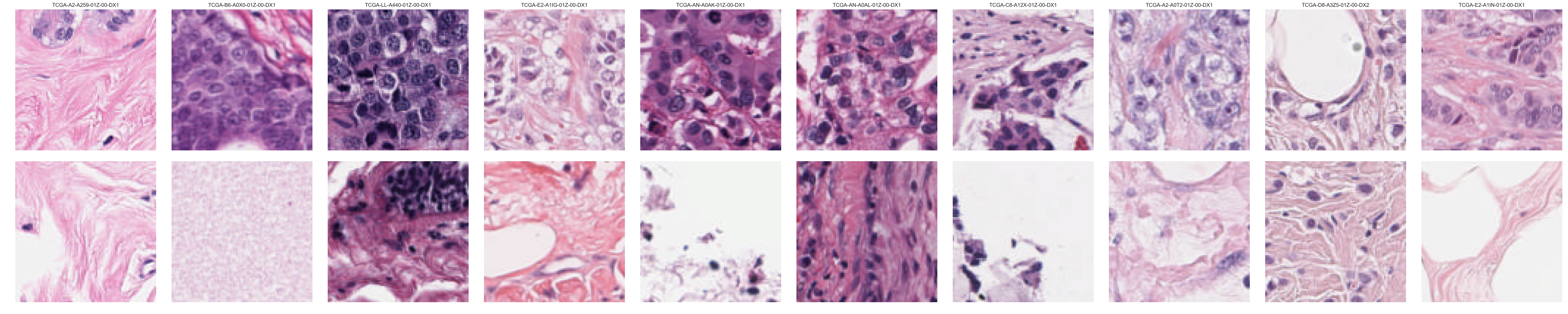
The image above shows the classification result of the NSANet classifier. The first line shows the areas classified as useful, and the areas classified as non-useful are in the second line. As you can see, the distinction is evident.
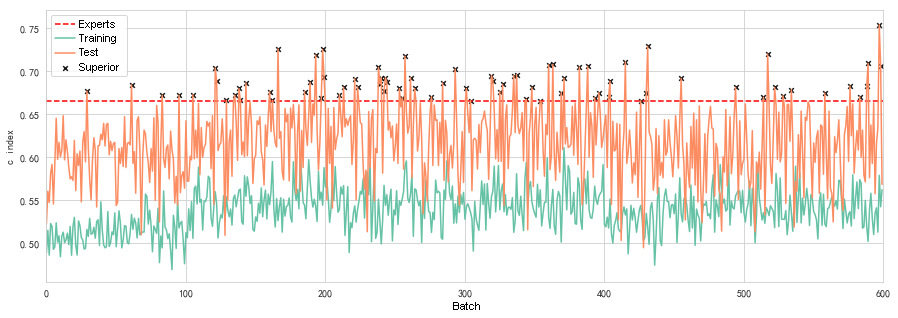
Compared to the survival analysis model trained on experts’ extraction features, this model can achieve similar performance. The models with the red marker are which with better performance than the base Cox PH model.
There are 78 better than the Cox HP basic model obtained under the same conditions. The records of the performance of 600 models in the figure above show that the performance changes are random. Because each training randomly selects a region from the samples, sometimes the area contains valid information, and sometimes the region does not have valid information. Therefore, although each model’s parameter training is based on the previous model, the latter The trained model is not necessarily better than the previous one. In general, the probability that a model is better than the Cox HP basic model is 13%. If we want 1−(1−0.13)^n ≤ 0.999, we can get n ≥ 50, which means training for 50 times or more, the probability of getting a model with a predictive power better than the Cox HP basic model is higher than 99.9 %.
For one case, the randomly selected useful areas are marked in the image below. These green boxes mark them.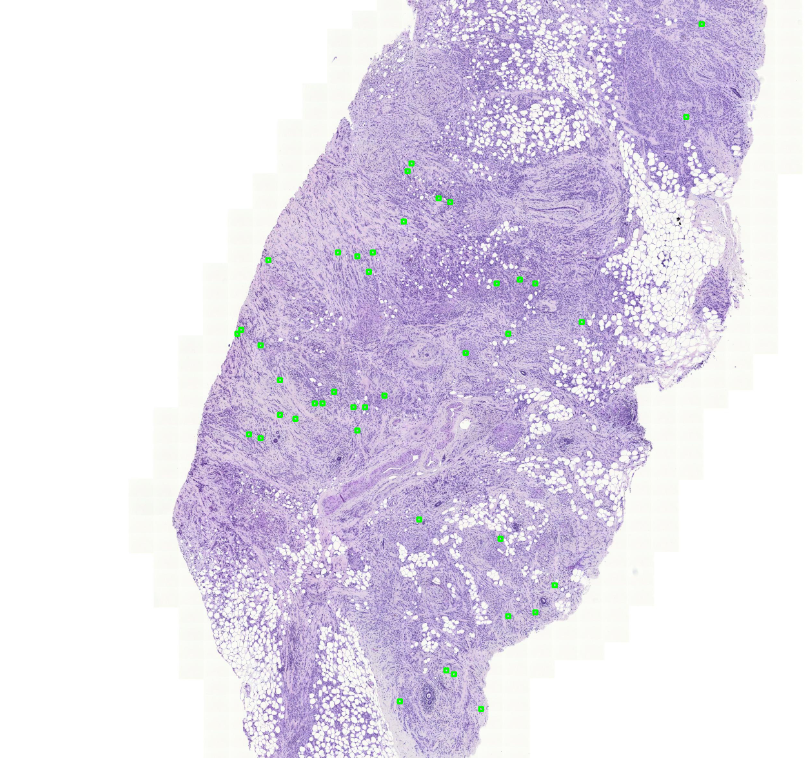
If we pore over these areas, we get the images below where the individual areas under 10X power imaging are gathered together.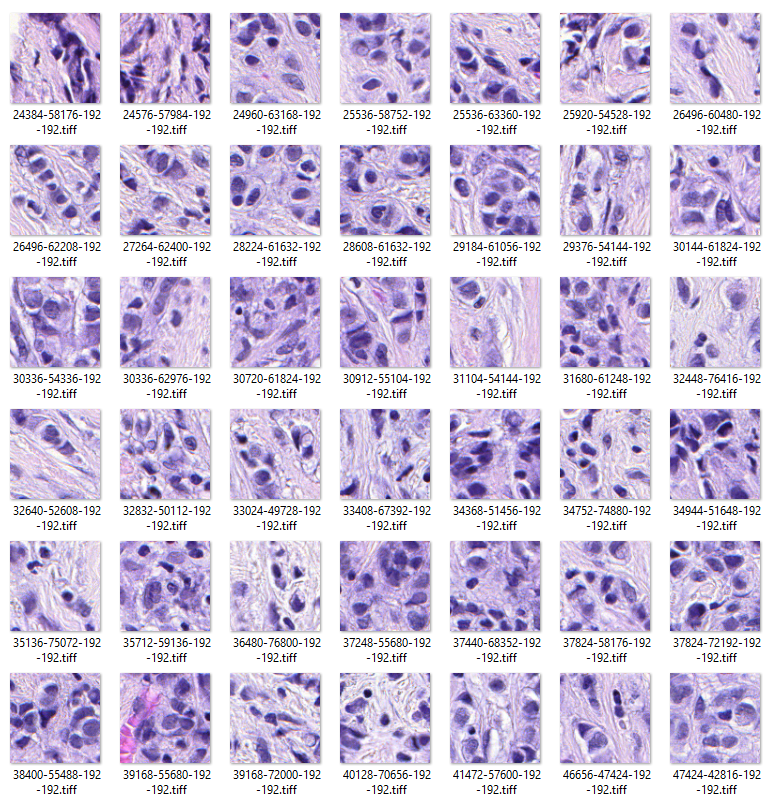
The model can be fully automately trained from the .svs pathological slice images. The model is also medium in size, with 7.24e+07 parameters, of which 4.26e+07 parameters are frozen from the model pre-trained on Kaggle data. Hence, the actual number of parameters to be calculated is only 2.97e+07.




