Introduction
Magnetic resonance imaging (MRI) is a medical imaging technique used to form pictures of the body’s anatomy. It can be applied to scan the brain to reveal the changes inside. For edema, it is recognized in MRI (T2 weighted or FLAIR pulse series) as a bright signal. Using the computer vision (CV) technique, we can extract and quantify the bright signals. Since these signals reflect each patients’ edema’s traits, doctors may rely on this information to better understand one’s edema’s idiosyncrasy and make suitable decisions. So we, the researchers in Sun Yat-sen Memorial Hospital started this project to study whether the outputs from MRI CV analysis can help doctors to decide which therapy fits the patients’ best interests.
We collected coronal 132 series of T2-flair Digital Imaging and Communications in Medicine (DICOM) images from 66 patients before and after their first therapy. My task is to write a program that can extract the bright areas from these images and quantify the features contained within them.
Main Problems
How to segment the bright areas from DICOM images?
How to reconstruct 3D edema models to show the result of segmentation?
How to extract features from these regions of interest?
Solutions I applied
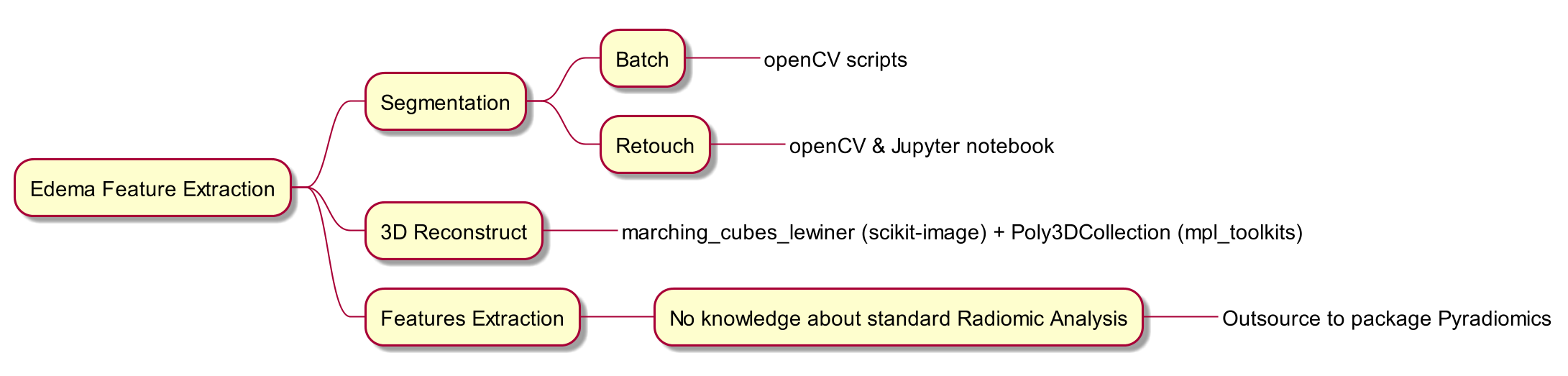
Resize images to 320 * 320, record changes in Spacing
Segment edema areas via OpenCV.
Calculate volume changes, quantify features via PyRadiomics.
Select features related to “Shape”, “ASM”, “Contrast”,
“Correlation”, “Homogeneity”, “Entropy”, “Variance”,
“Skewness”, “Kurtosis” by T-test.
See more details in codes.
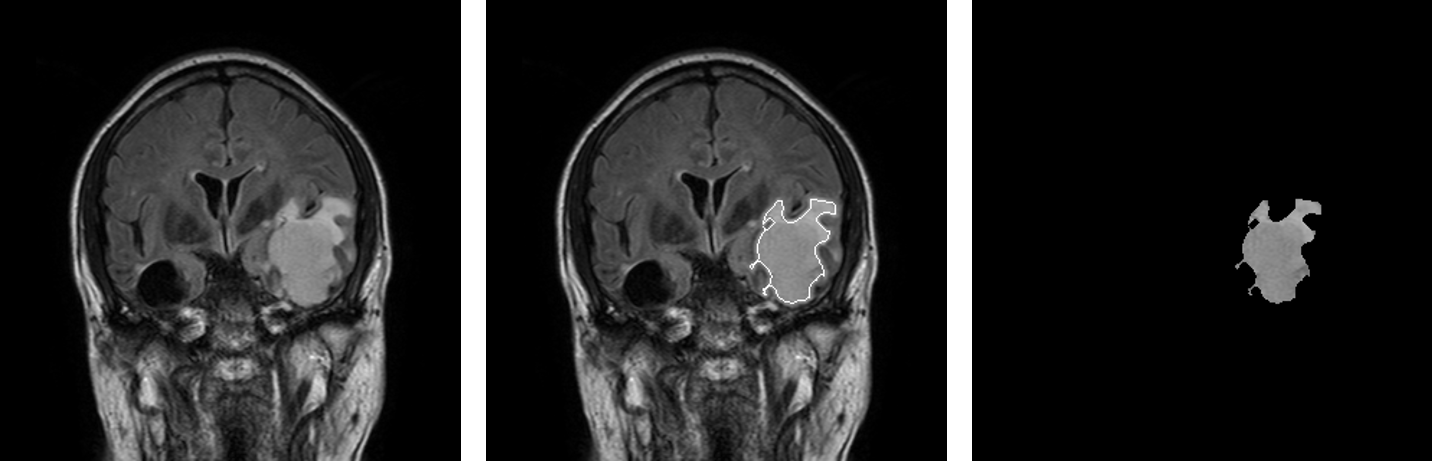
Results
The result of segmentation
The result of Reconstruct





